Applications | Biotechnology: NMR Metabolomics
Unravelling the metabolomes of plants
Introduction
Nuclear Magnetic Resonance (NMR) methods have developed rapidly during the past 20 years, enabling the study of molecular identity, composition and fluxes in surprising new ways and (live) materials. Even though mass spectrometry (MS) using uniformly labelled 13C plant materials is becoming a dominant tool in (non-)targeted metabolomics (e.g. Kikuchi et al, 2004; Pinta et al, 2018; Tsugawa et al, 2019), NMR has its advantages in the overall qualitative and quantitative characterization of metabolites, especially when combined with stable isotope enrichment. NMR and MR-imaging (MRI) are at the center of exciting, novel applications. Proton, 13C, and/or 15N NMR of “crude” plant extracts provide us with a fingerprint of a wide range of extractable metabolites (e.g. Kikuchi et al, 2004; Nakabayashi and Saito, 2017; Van Erven et al, 2019).
The ability to detect metabolites without complex sample preparations makes NMR suitable for in vivo and high-throughput analyses like the screening of large numbers of mutants versus wildtypes (Fukushima et al, 2002; Kikuchi et al, 2004), especially when combined with mass spectrometry (Nakabayashi and Saito, 2017; Van Erven et al, 2019) and nowadays even in vivo in developing plants (Comprehensive Multi Phase (CMP)-NMR, e.g. Fortier-McGill et al, 2017), in new organic solar cells for hydrogen production (Wakerley et al, 2017), and in vivo in human cancer tissues (Dual-Agent Hyperpolarized 13C MRSI, Bok et al, 2019). However, a drawback of NMR is its low sensitivity with natural biomass, especially with 13C and 15N.
Stable Isotope solution: Enrichment for NMR
The low sensitivity has been overcome with stable isotope enrichment and new NMR methods. Enrichment with stable isotopes like 13C and 15N enables identification and quantification of metabolites, and ultimately of complete metabolomes, by combining uniformly labelled metabolites with various NMR and MS techniques. A basic concept of NMR-based plant metabolomics has been proposed by Kikuchi et al (2004). These authors produced uniformly labeled Arabidopsis plants in tissue culture by feeding 13C-labelled glucose and 15N-labelled nitrate as the only C- and N- source, respectively. Stable isotopes like 13C and 15N enable scientists in metabolomics to identify the metabolites of complete metabolomes by combining uniformly labelled metabolites with NMR. For example, in this way the differences in the metabolomes of the wildtype and mutants of Arabidopsis have been studied (Kikuchi et al, 2004). By labelling plants with the stable isotopes 13C and/or 15N during a complete life cycle from seed to maturity, all organs and plant components will contain the isotopes in virtually the same ratio as present in the substrate. When the substrates contain 99% 13C and/or 99% 15N, all plant components will be uniformly labelled at nearly the same 99% enrichment. Subsequently, the stable-isotope labelled components of interest (biomass, fibers, or metabolites) can be extracted and mixed with samples of non-labelled extracts for analysis by NMR with or without MS.
Figure 1 illustrates how plants were uniformly labelled with the stable isotopes 13C and 15N (1) and subsequently analysed with hetero-nuclear 2D NMR (2). The stable isotope labeled plants showed a high-level and a high-sensitive separation of metabolite signals (3). Subtraction of spectra (4) of different mutants yielded a quantitative figure of increased and decreased metabolites (5) in the samples (Kikuchi et al, 2004).

Figure 1. Principle of comparative metabolite analysis with NMR (Kikuchi et al, 2004).
In 2004, the authors concluded that NMR will become a key technology in plant metabolomics: “the use of isotope labeling together with newly developed NMR technologies opens a new avenue for plant metabolomics” (Kikuchi et al, 2004).
Stable Isotope solution: Photoreforming of (13C) cellulose to H2 using CdS/CdOx
In Nature Energy, Wakerley et al (2017) demonstrated a remarkable discovery, the photocatalytic production of H2 from cellulose in the presence of a catalyst by the identification of products of U-13C alpha-cellulose oxidation by 13C-NMR spectroscopy using IsoLife’s uniformly 13C-labelled cellulose. Figure 2 shows the formation of three 13C-labelled products originating from 13C-labelled cellulose photo-oxidation in the presence of a CdS/CdOx catalyst: carboxylic acid (signal ii), formate (iii), and carbonate (iv).
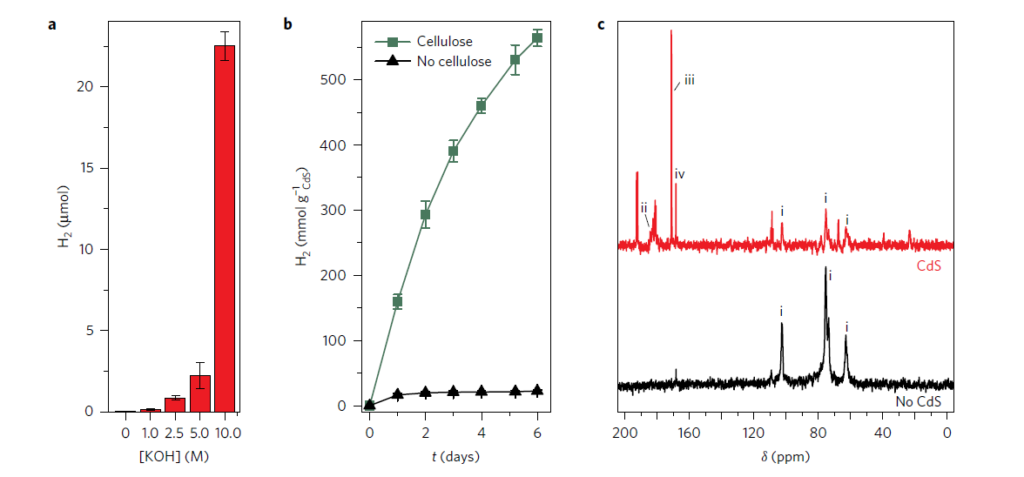
Figure 2. Formation of three 13C-labelled products (Wakerley et al, 2017).
Stable Isotope solution: Enrichment for CMP-NMR on live, germinating 13C seeds
Comprehensive Multi Phase (CMP)-NMR has been used for simultaneous monitoring in the liquid, gel and solid state of metabolic changes in germinating uniformly 13C-enriched wheat seeds in the NMR machine probe (Lam et al, 2014; Fortier-McGill et al. 2017). Figure 3 shows the percentage change of both the anomeric carbohydrate signals (93−103 ppm) and the TAG (triacylglycerides, 125−137 ppm) and from the edited spectra: IDE (dissolved small freely diffusing), DE (rigid gels semisolids with restrictive diffusion), RADE (large fast relaxing semisolids), at time 24, 48, 72, and 96 h relative to the dry seed at 0 h (Fortier-McGill et al, 2017).
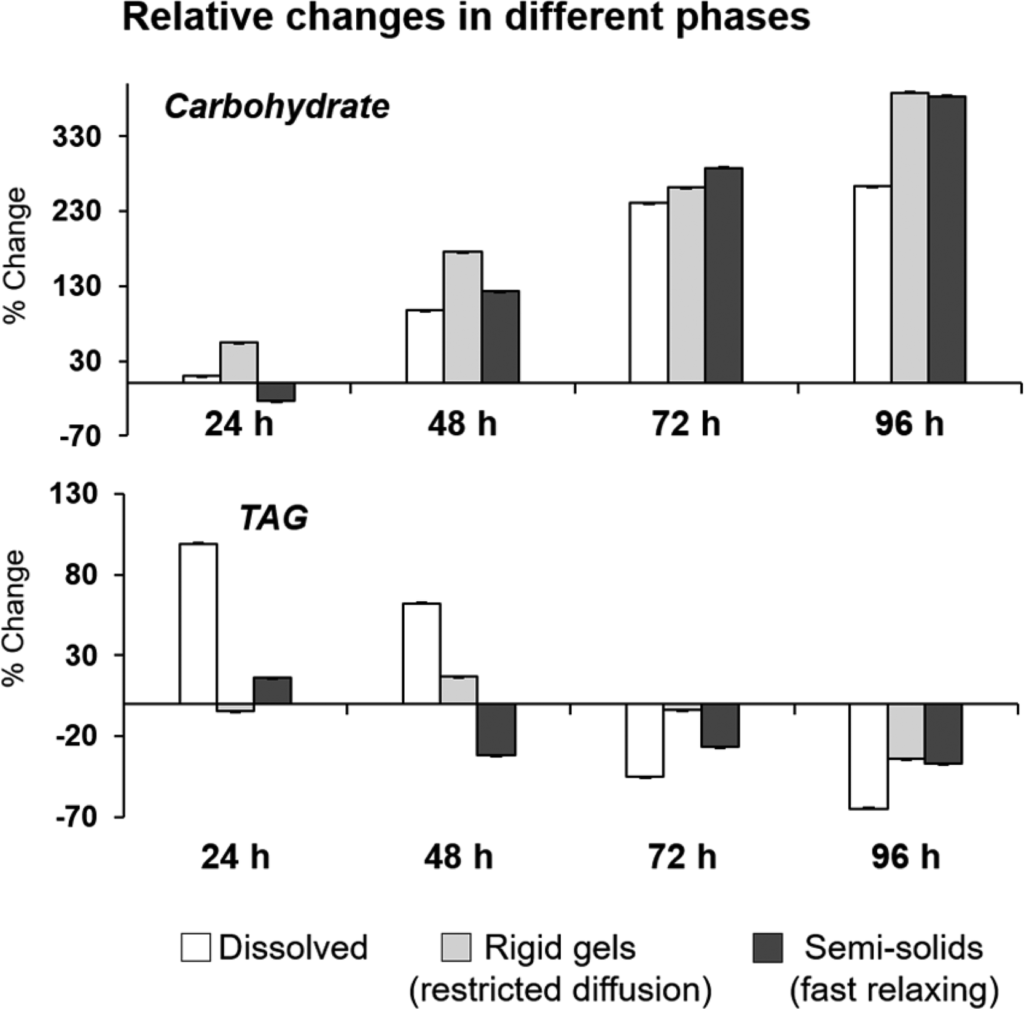
Figure 3. Relative changes in carbohydrate during germination of U-13C wheat seeds using CMP-NMR (Fortier-McGill et al, 2017).
References
Bok R, J Lee, R Sriram, K Keshari, S Sukumar, S Daneshmandi, DE Korenchan, RR Flavell, DB Vigneron, J Kurhanewicz, P Seth. 2019.
The role of lactate metabolism in prostate cancer progression and metastases revealed by dual-agent hyperpolarized 13C MRSI.
Cancers 11: 257.
Fortier-McGill B, RD Majumdar, L Lam, R Soong, YL Mobarhan, A Sutrisno, R de Visser, MJ Simpson, H Wheeler, M Campbell, A Gorissen, AJ Simpson. 2017.
Comprehensive multiphase (CMP) NMR monitoring of the structural changes and molecular flux within a growing seed.
Journal of Agricultural and Food Chemistry 65: 6779–6788.
Fukushima K, J Kikuchi, S Koshiba, Y Kuroda, S Yokoyama. 2002.
Solution structure of the DFF-C domain of DFF45/ICAD. A structural basis for the regulation of apoptotic DNA fragmentation.
Journal of Moleculair Biology 321: 317–326.
Kikuchi J, K Shinozaki, T Hirayama. 2004.
Stable Isotope Labeling of Arabidopsis thaliana for an NMR-Based Metabolomics Approach.
Plant Cell Physiology 45: 1099–1104.
Lam L, R Soong, A Sutrisno, R de Visser, MJ Simpson, H Wheeler, M Campbell, WE Maas, M Fey, A Gorissen, H Hutchins, B Andrew, JO Struppe, S Krishnamurthy, R Kumar, M Monette, H Stronks, A Hume, AJ Simpson. 2014.
Comprehensive multiphase NMR spectroscopy of intact 13C labeled seeds.
Journal of Agricultural and Food Chemistry 62: 407-415.
Nakabayashi R, K Saito. 2017.
Ultrahigh resolution metabolomics for S-containing metabolites.
Current Opinion in Biotechnology 43: 8-16.
Pinta MN, I Montoliu, A-M Aura, T Seppänen-Laakso, D Barron, S Moco. 2018.
In vitro gut metabolism of [U-13C]-quinic acid, the other hydrolysis product of chlorogenic acid.
Molecular Nutrition & Food Research 62: 1800396.
Tsugawa H, R Nakabayashi, T Mori, Y Yamada, M Takahashi, A Rai, R Sugiyama, H Yamamoto, T Nakaya, M Yamazaki, R Kooke, JA Bac-Molenaar, N Oztolan-Erol, JJB Keurentjes, M Arita , K Saito. 2019.
A cheminformatics approach to characterize metabolomes in stable-isotope-labeled organisms.
NATURE Methods 16: 295-298.
Van Erven G, R de Visser, P de Waard, WJH van Berkel, MA Kabel. 2019.
Uniformly 13C labeled lignin internal standards for quantitative pyrolysis−GC−MS analysis of grass and wood.
ACS Sustainable Chemistry & Engineering 7: 20070−20076.
Wakerley DW, MF Kuehnel, KL Orchard, KH Ly, TE Rosser, E Reisner. 2017.
Solar-driven reforming of lignocellulose to H2 with a CdS/CdOx photocatalyst.
NATURE Energy 2: 17021.